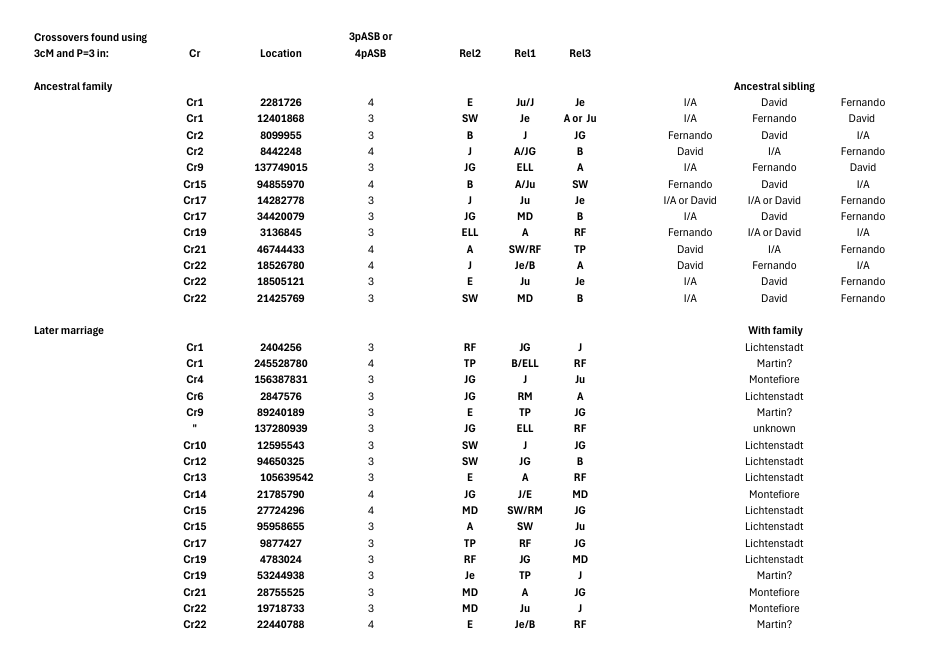
A
discovery was made which from the outset seemed promising but a little hard to
understand. To
explain, 'abutting
segment boundaries' or ASBs are a type of segment boundary
coincidence in which 2 segment matches abut (we first reported
2 such cases
here in which the Cr2 8099555 event is accompanied by
the additional 8442248 event). Initially we (mistakenly) saw an
explanation of ASBs in meiosis, for 4 gametes are produced from one set of parental chromosomes. Complementary gametes show a parent's DNA rejected on one side of a
recombination appearing on the other side, thus producing an abutting boundary
coincidence which can survive to later generations. With some excitement our presentation
'Fun with Autosomal DNA' used this 'insight' to claim a Baruch
Lousada connection across 4 family branches. However Andrew Millard pointed out
that the statistics of sperm and egg utilization do not favour our 'insight'.
After our first 2 ASBs, new ones were discovered.
From what follows, it seemed that the terminal SNP of any reported
segment normally lies outside the actual segment.
For as GEDmatch advised on 24 Oct 2025: 'The
boundaries of a segment are practically impossible to get exactly correct.
There are alleles in positions which are not SNPs and the crossover is
likely not exactly at a SNP. So the mismatch at the front of the segment is
before the segment actually starts and the mismatch after the end of the
segment is after the segment ends. There is also the possibility that the
SNPs after the beginning of the mismatch at the beginning of the segment
happen by chance and the segment might actually start after the first
aligned SNP following the match. For example if the first SNP inside the
segment is AC then it will match what ever is in the other kit but the
crossover may be further in. The same is true at the end of the segment.
Finally if the two kits are not from the same vendor chip set there are SNPs
which do not align and had those SNPs been available the mismatch SNPs which
bound the segment may be different'.
However, GEDmatch later advised that it treats opposite ends of a segment
differently - including the first non-matching SNP at one end and using the
last SNP before a mismatch at the other. This explains why ASBs are not reported as having a
1 SNP overlap, for the same SNP will be reported as the end of both segments
- being included in one segment and outside the other - and defines the ASB. Qmatch was
recommended to us by GEDmatch for use with small matches, and we have indeed found
that without it ASBs do not appear. In Qmatch while it was easy
for us to notice some ASBs unaided, because the end of one segment is reported as
numerically identical to the beginning of the other abutting segment, AI will be needed to
avoid missing any ASBs, and this would also allow a search for ASBs showing
misses of a small number of nucleotide positions. In fact, even with our 13 person sets, AI
showed that ASBs were missed (4 out of 46 for the comparison set of 13 plus
an error identification, and
9 out of 31 for the 13 relatives). However, a note of caution is appropriate
for an ASB while representing crossovers can also represent
pseudo-crossovers. This is because another segment terminating anywhere
between the other first included SNP and the ASB will also show up at the
ASB. We plan to use AI to gauge the likelihood of pseudo-crossovers
represented at ASBs for both relatives and randoms.
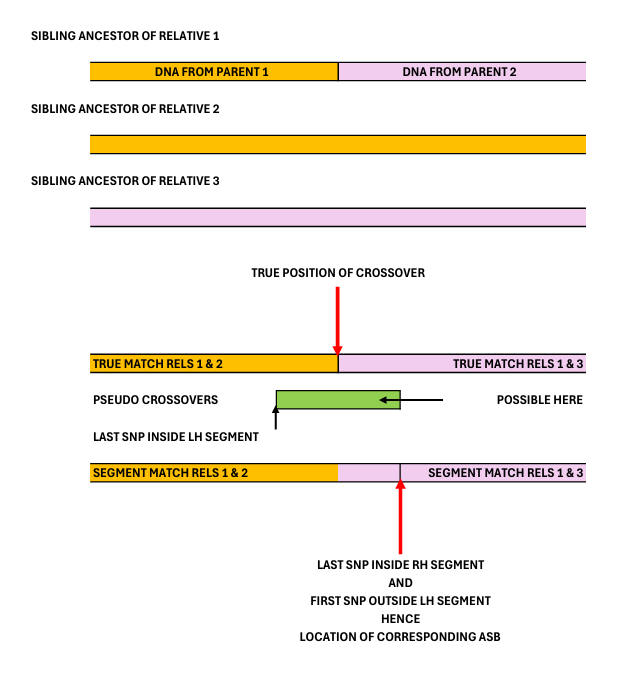
The following diagram shows in essence what causes ASBs - starting with
3-person ASBs (3pASBs).
Thus, where an ancestral crossover in one
sibling is carried forward into a present-day descendant and is accompanied (in
2 further present-day descendants) by a stretch of each parent's DNA which
bridges the crossover, ASBs can result under
favourable circumstances. By this is meant that they will obviously not be detectable in cases where the ancestral parents match in the
region surrounding the crossover. It will be noted that the ASB generally
doesn't match the true crossover position, and that in the region in green
pseudo-crossovers are possible. In addition, the 2 SNPs defining the green
region will only the 2 closest SNPs on either side of the true crossover if
both parents share them. In any event, it is important to consider
whether the ancestors defining an ASB are 'parents' as shown in the diagram
and not close relatives thereof:
In any case, our insight allows us to also understand
4-person
ASBs as well - for here, a pair of relatives from the same family branch
can act as a surrogate for Relative 1 in the above chart. That is, both of
the relatives in the pair carry the ancestral crossover.
We can now see the potential of ASBs, for in the following chart of proven
matches, we have added 31 extra ASB connections (detailed below). It can be seen that
in total these ASB contributions greatly outnumber the 5 matches
from RSBCs and the 10 from Qmatch.